Annotation Output¶
An interactive example is available at https://pnnl.github.io/hundo/.
OTU.biom
Biom table with raw counts per sample and their associated taxonomic assignment formatted to be compatible with downstream tools like phyloseq.
OTU.fasta
Representative DNA sequences of each OTU.
OTU.tree
Newick tree representation of aligned OTU sequences.
OTU.txt
Tab-delimited text table with columns OTU ID, a column for each sample, and taxonomy assignment in the final column as a comma delimited list.
OTU_aligned.fasta
OTU sequences after alignment using MAFFT.
all-sequences.fasta
Quality-controlled, dereplicated DNA sequences of all samples. The header of each record identifies the sample of origin and the count resulting from dereplication.
blast-hits.txt
The BLAST assignments per OTU sequence.
summary.html
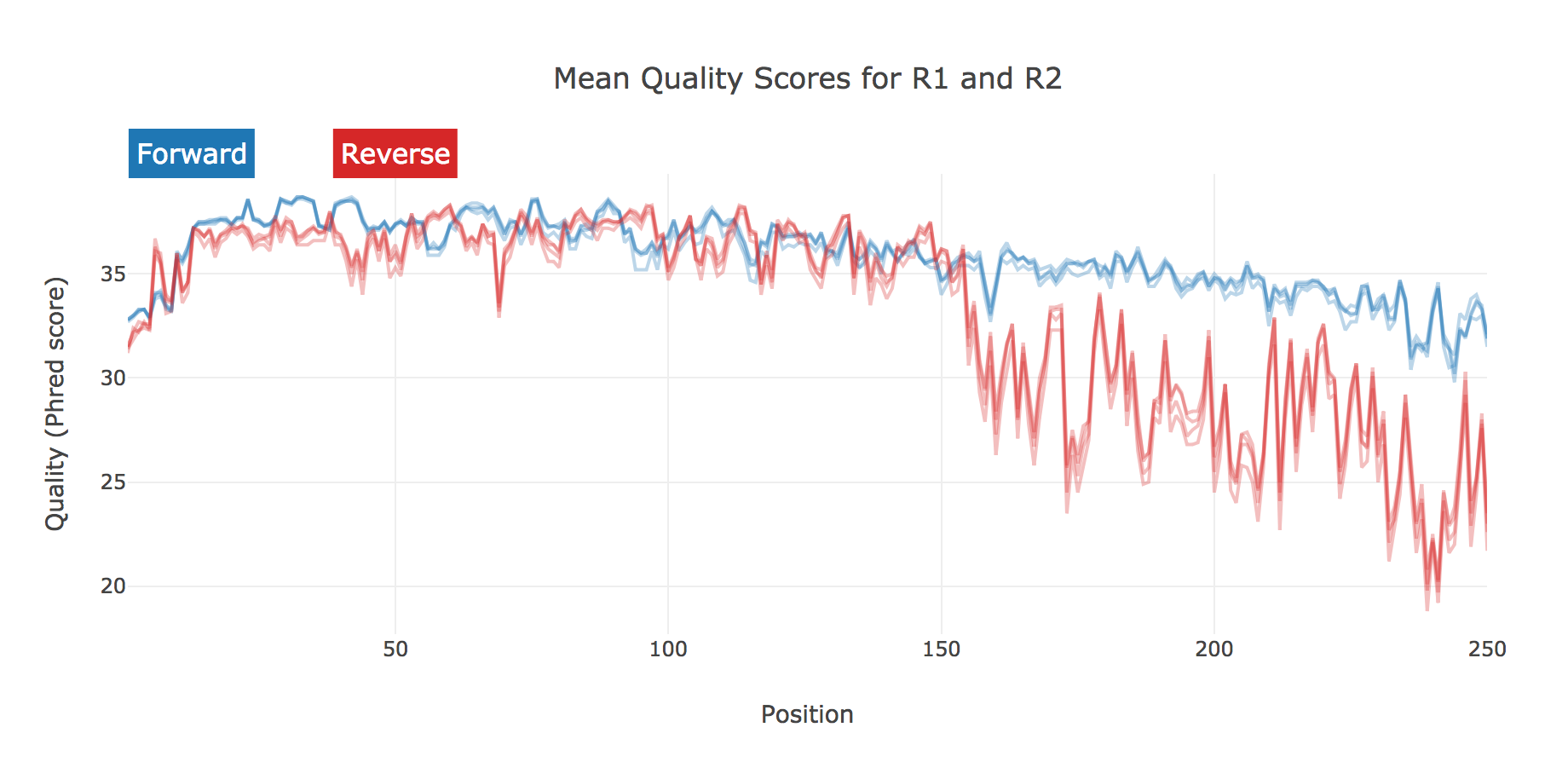
Captures and summarizes data of the experimental dataset. Things like sequence quality:
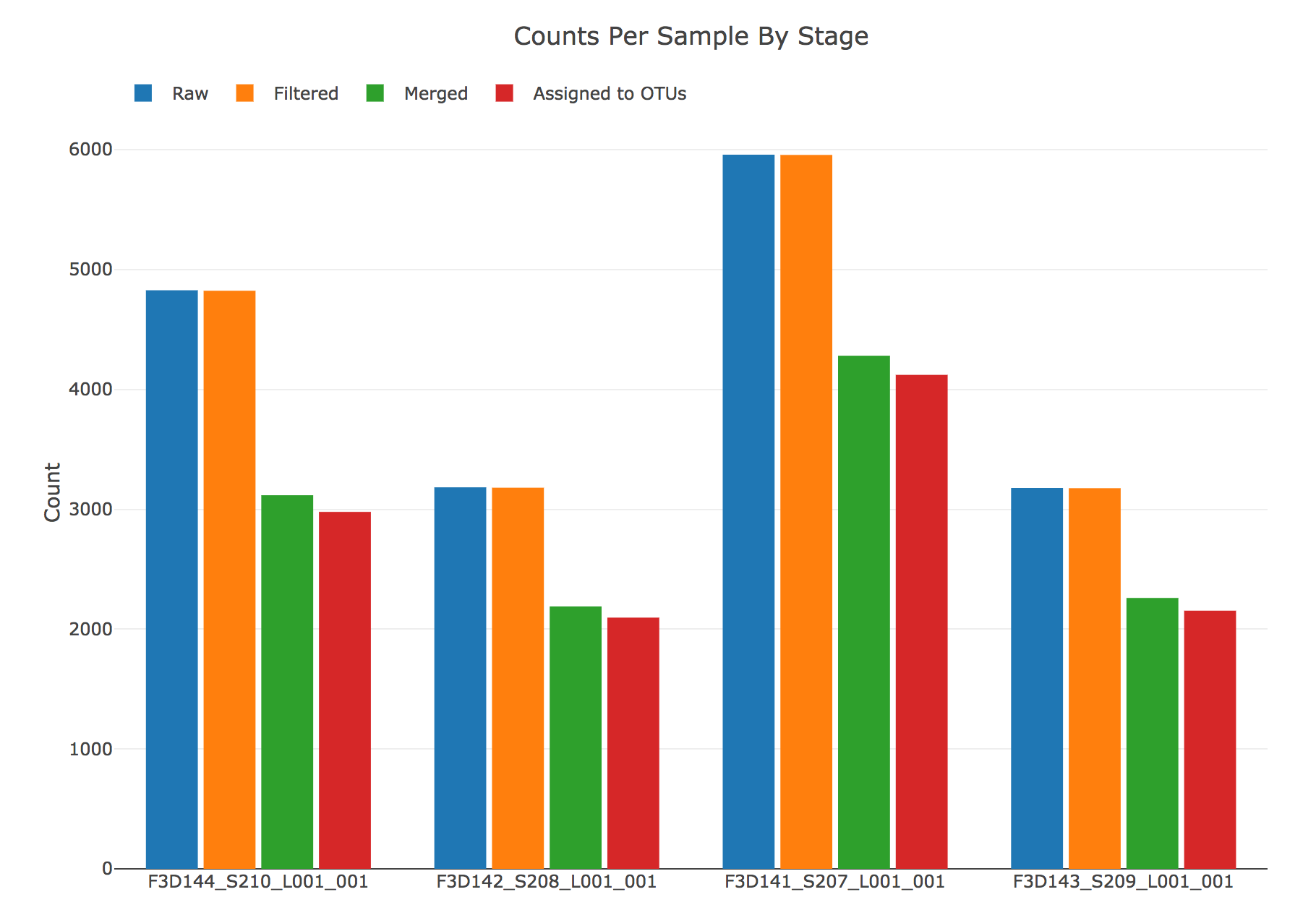
And counts per sample at varying stages of pre-processing:
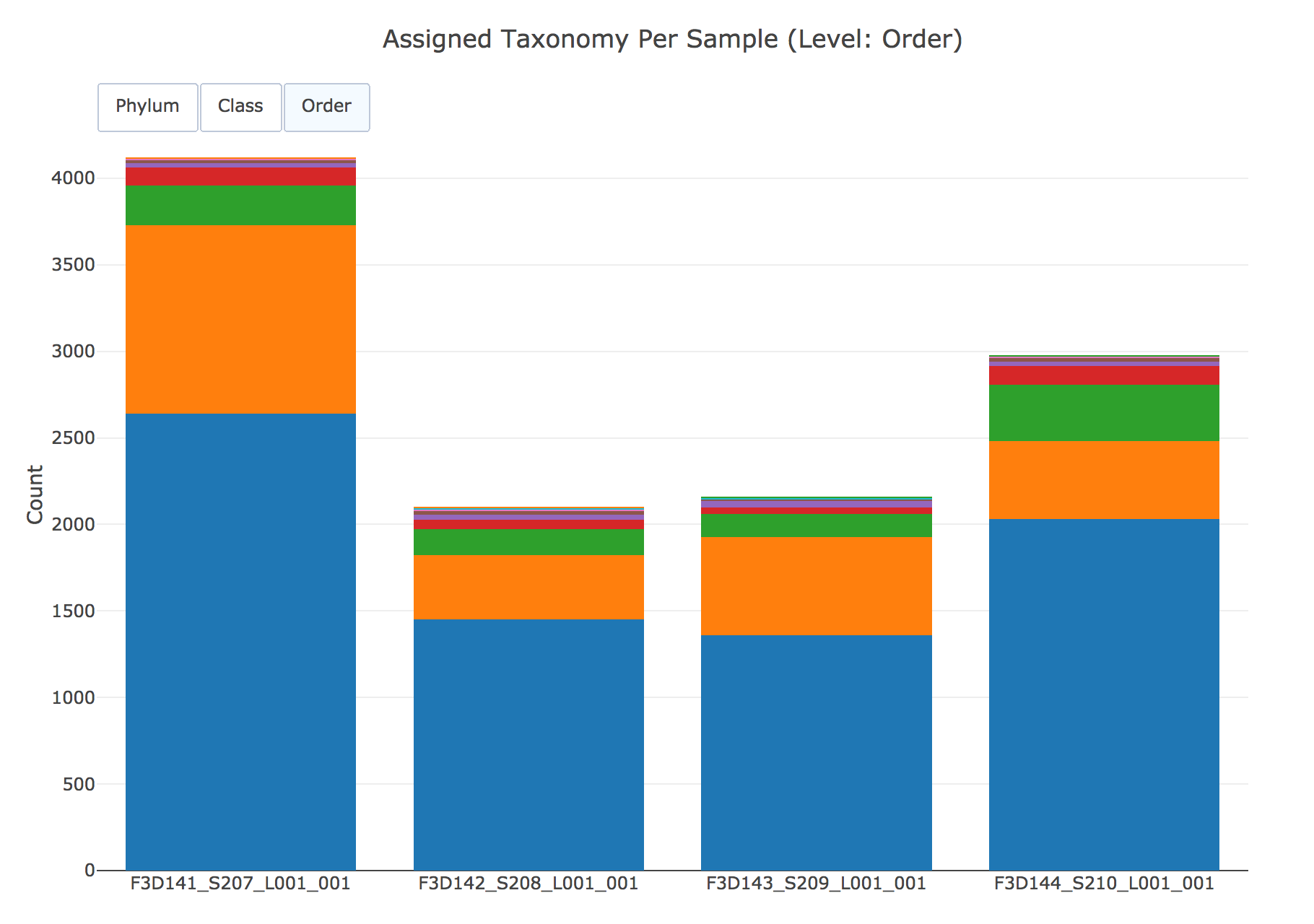
Taxonomies are also summarized per sample across phylum, class, and order: